Based on an article in Nature Chemical Biology by Tristan de Rond, Julia Asay, and Bradley Moore.
This tool is a work-in-progress. Please raise issues or ask questions in the discussion group.
Code for the Jupyter Notebook version is available at github.com/tderond/CO-ED.
This tool generates files that need to be opened in Cytoscape, which is free open source software. (Instructions)
The CO-ED networks generated by this tool need to be opened in Cytoscape, which is free open source software that can be downloaded from www.cytoscape.org
The tool generates files for undirected and directed versions of the network.
The undirected network has one edge per co-occurring pair of domains, while the directed network has two (one in each direction). The undirected networks are easier to navigate, however if you want to know what fraction of proteins with domain x also contain domain y, you will need the directed network.
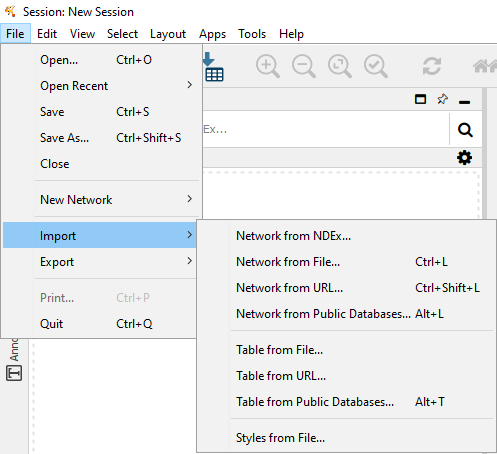
(Open one of these network files in Cytoscape under "File->Import->Network from File")
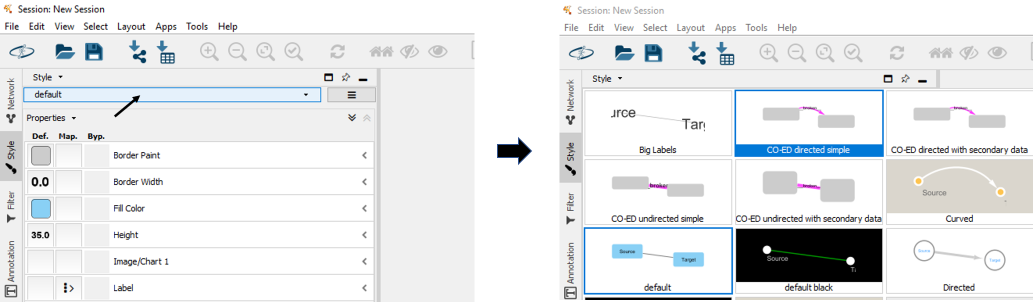
The style file allows you to visualize networks with formatting similar to that in the CO-ED manuscript. The style file is the same for all networks, no need to download it each time you generate a new network.
(Open the style file in Cytoscape under "File->Import->Styles from File", then select one of the CO-ED styles in the Style tab)
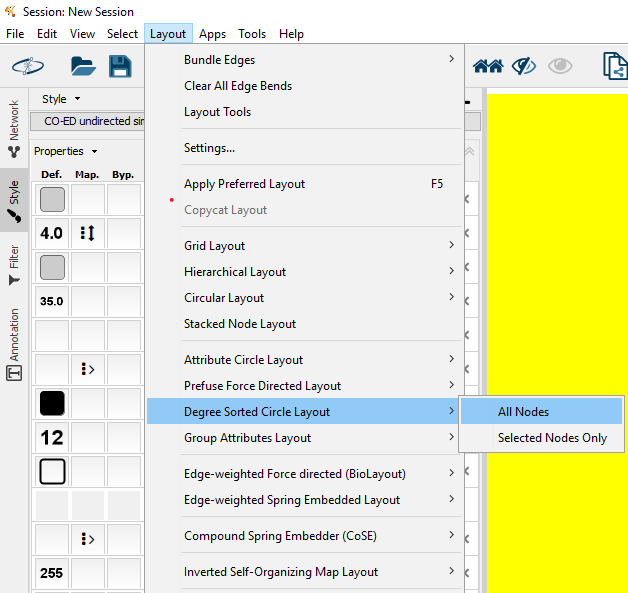
Initially when you open the network, all nodes will be overlapping. To spread the nodes out, apply a "layout" to the network. I recommend trying the "Degree Sorted Circle Layout" or "Hierarchical Layout"